Bayesian bi-clustering of categorical data
Alberto Pessia
Department of Mathematics and Statistics, University of Helsinki
European Meeting of Statisticians
Helsinki, 24-28 July 2017
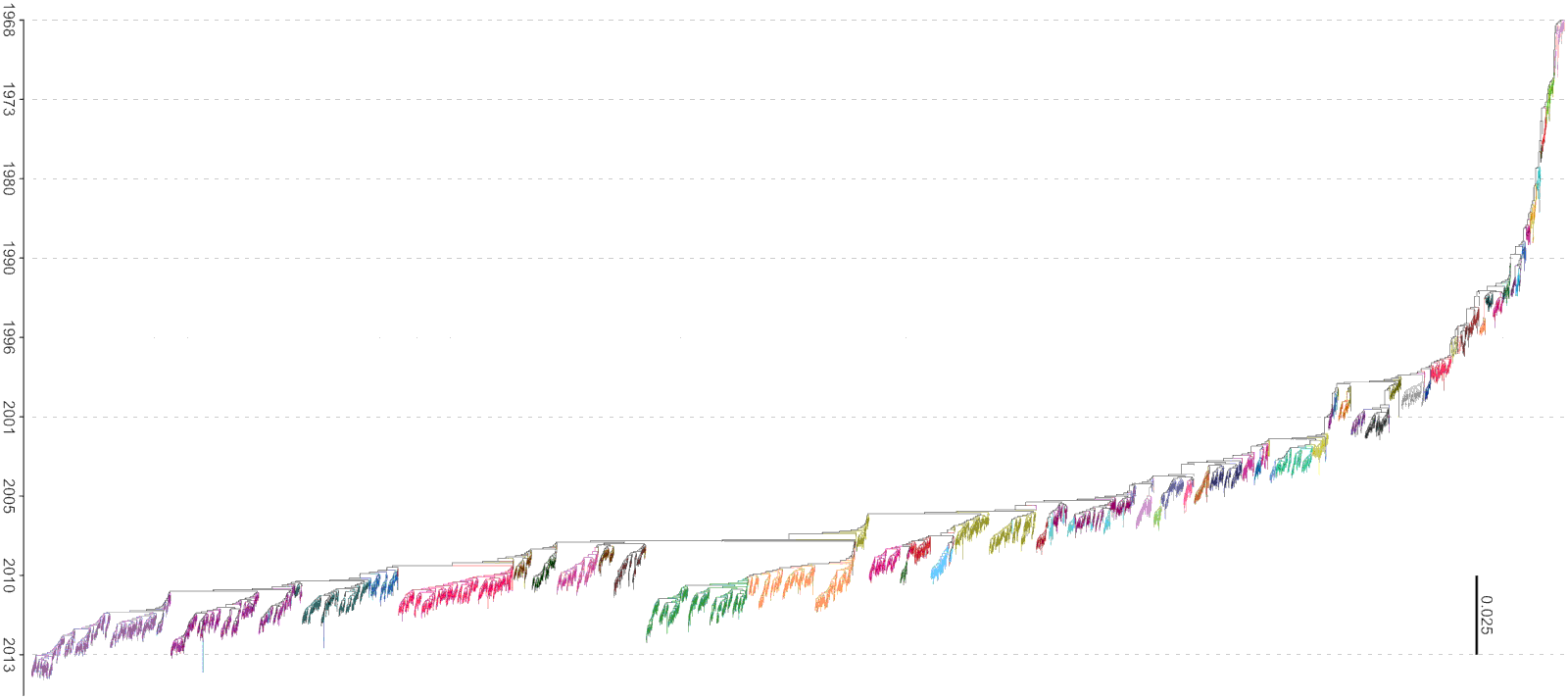
Motivation: identify groups of homogeneous proteins
Clusters naturally arise as a consequence of evolutionary processes
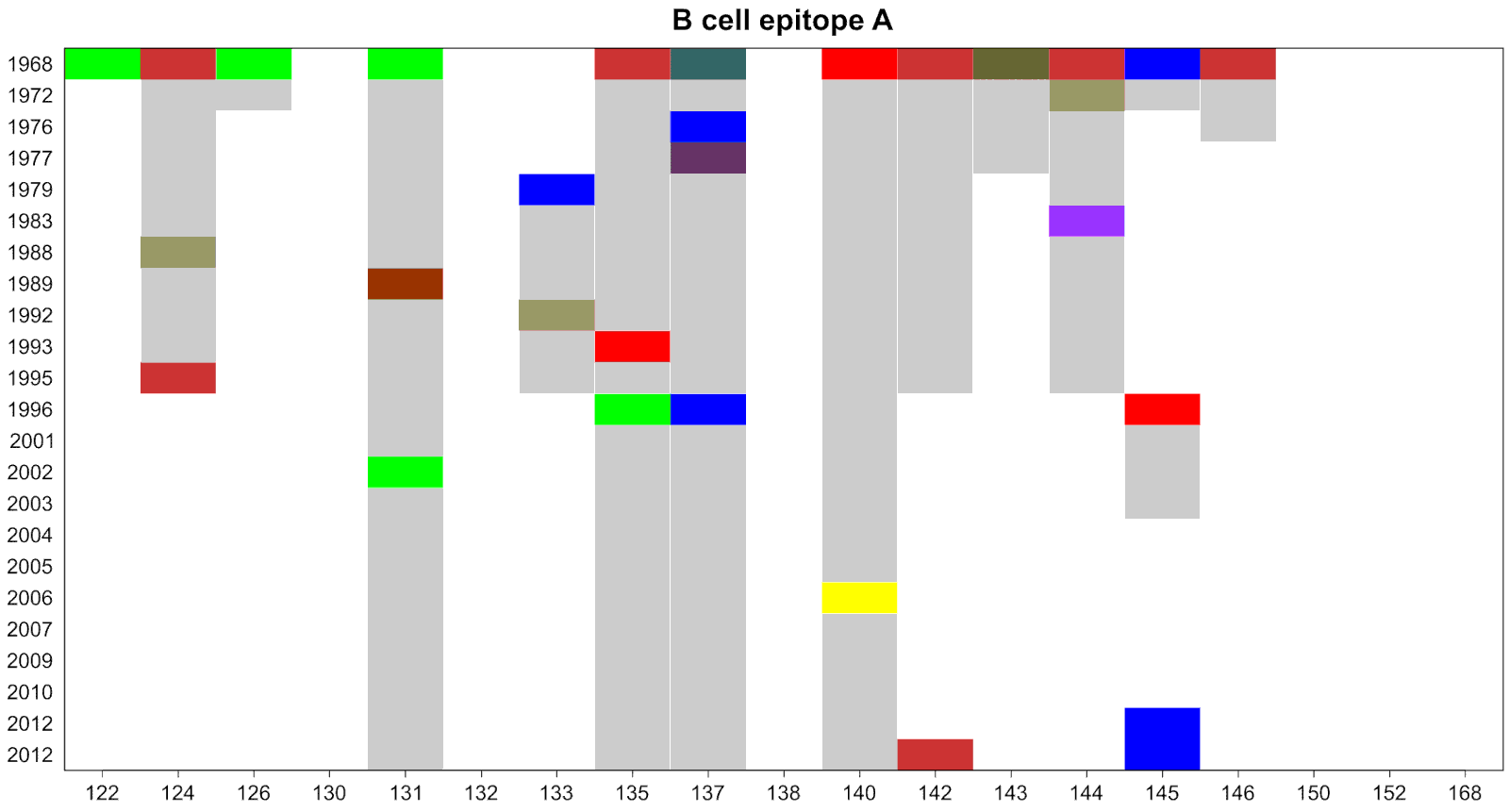
Motivation: identify important amino acids
Classify the sites and their corresponding amino acids according to their discriminatory power
Categorical data
No expected value, median, variance, etc.
Operations are done on counts and their probabilities
High dimensional
Usually thousands of rows and columns in the data matrix
Number of columns much greater than the number of rows
Bi-clustering
Partition the rows into blocks
Partition the columns into blocks as well
Bayesian analysis
Informative priors to regularize the likelihood
Posterior distribution to measure our uncertainty
Computation
Stochastic optimization to find the MAP
MCMC algorithms to explore the posterior
High costs for large dataset
Cluster analysis of categorical data
Status: Noise
We just received a black ball. In which cluster would you put it?
Status: Weak signal
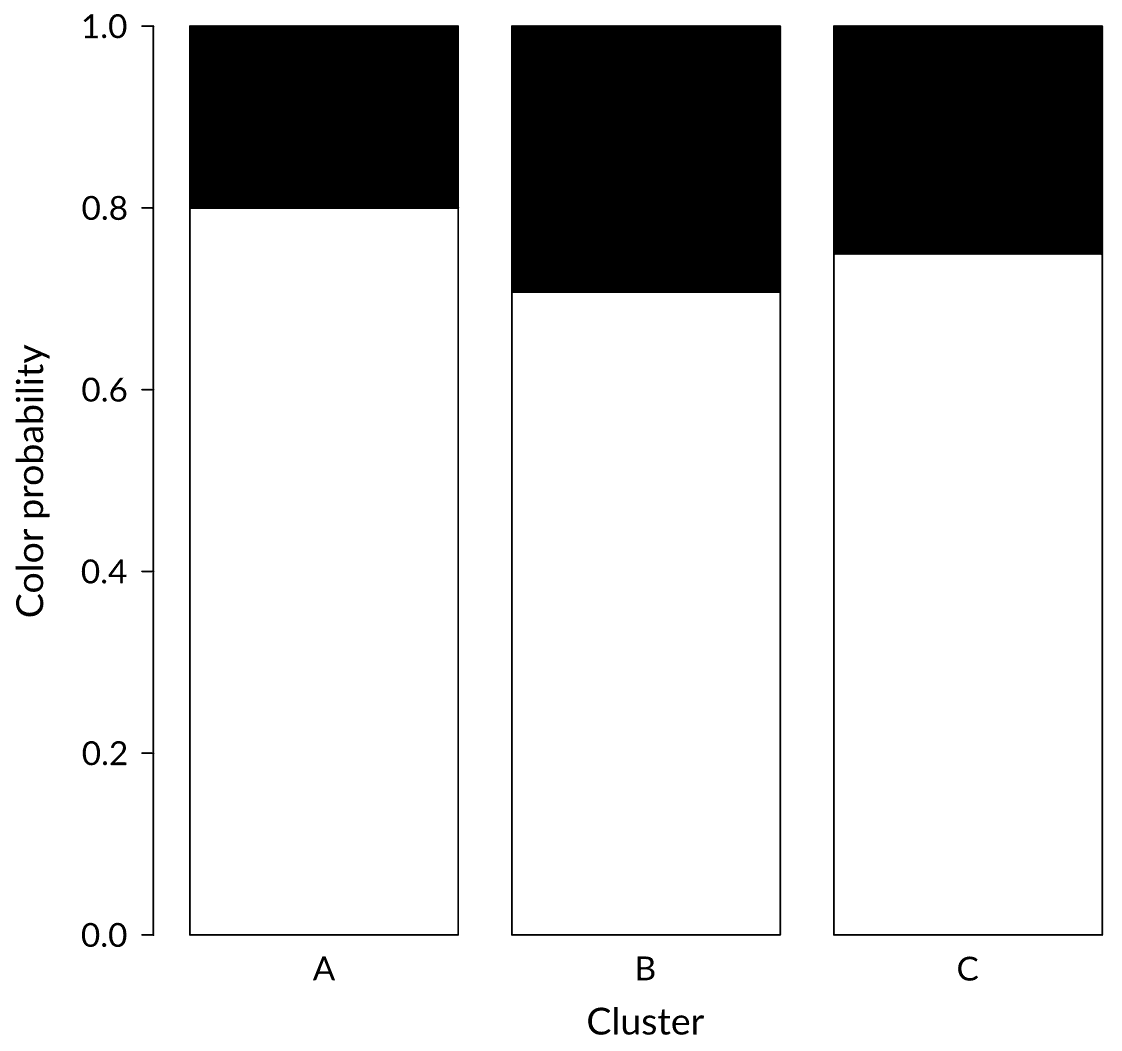
We just received a black ball. In which cluster would you put it?
Status: Strong signal
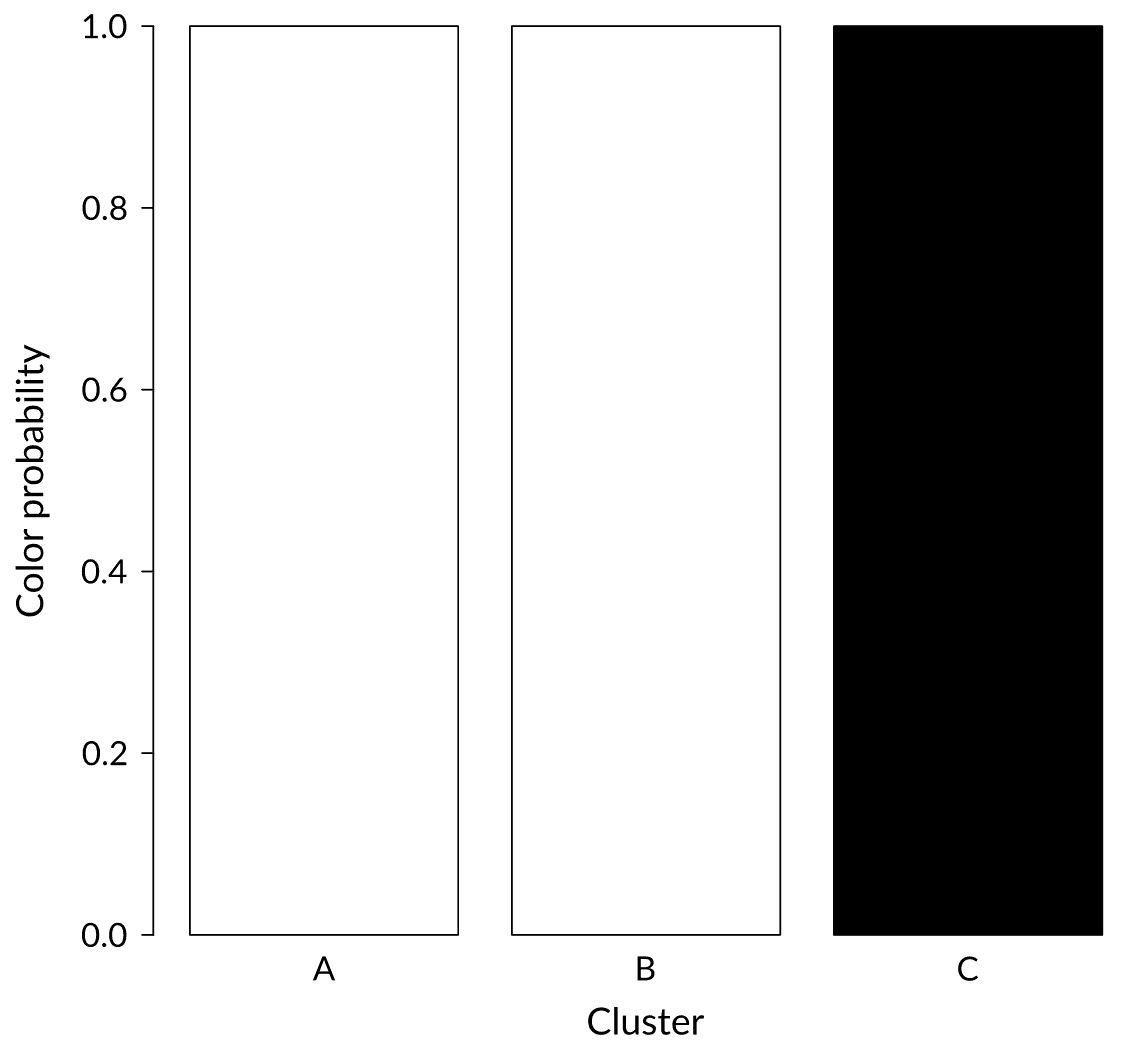
We just received a black ball. In which cluster would you put it?
Partitioned data matrix
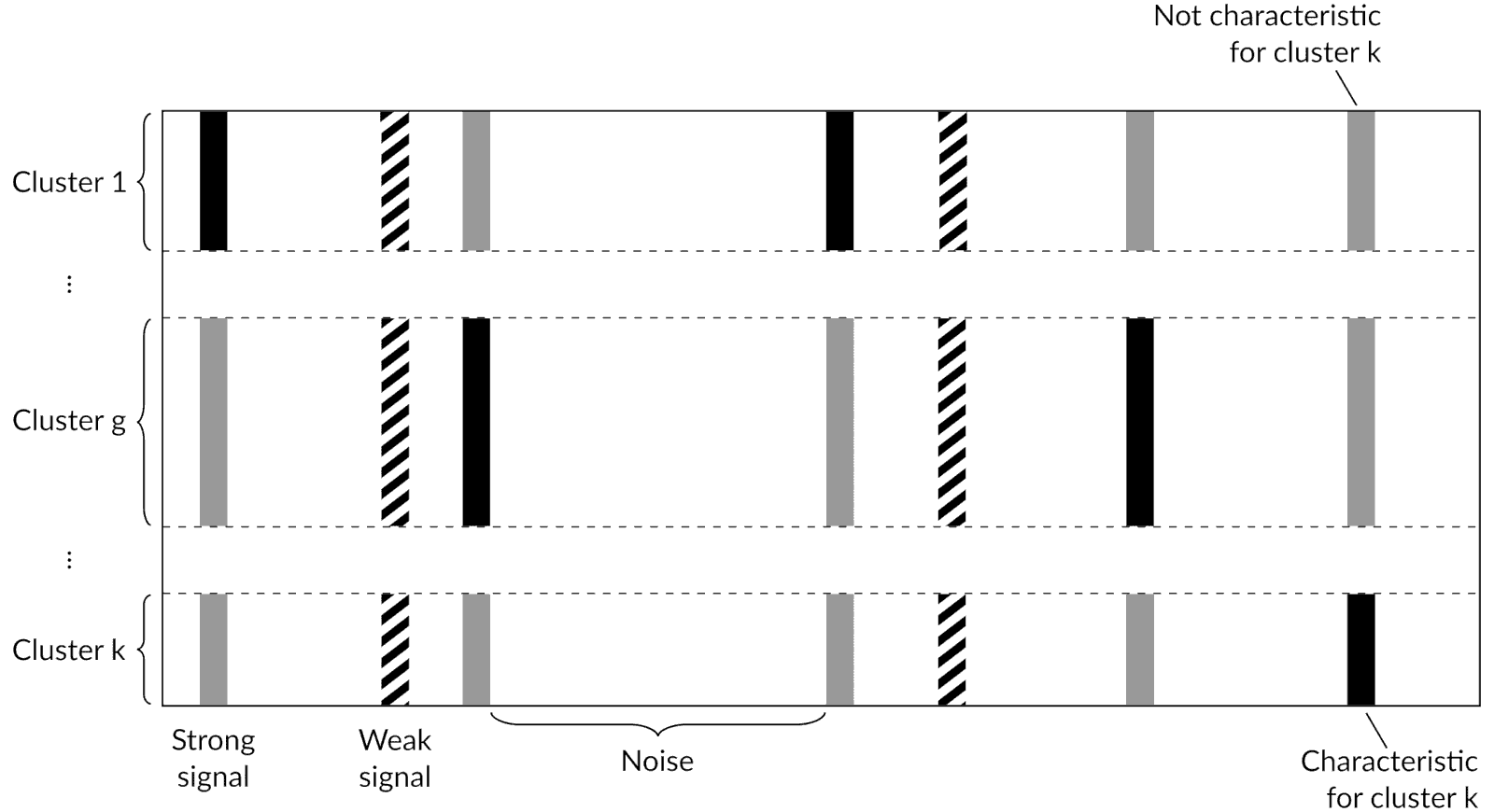
Model parameters
$k$ = total number of clusters
$R_{k}$ = partition of the rows into $k$ clusters
$C_{k}$ = partition of the columns among the $k$ clusters
where
$$
R_{kit} = \begin{cases}
1 & \text{if }i\text{ and }t\text{ belong to the same cluster}\\
0 & \text{otherwise}
\end{cases}
$$
$$
C_{kjgl}^{u} = \begin{cases}
1 & \text{if }j\text{ has status }u\text{ and property }l\text{ in cluster }g\\
0 & \text{otherwise}
\end{cases}
$$
Prior distribution on $R$
Model is flexible enough to accommodate any prior distribution on $(k, R_{k})$
In our implementation we used the Ewens-Pitman distribution
$$p(k, R_{k}) = \frac{\Gamma(\psi)}{\Gamma(\psi + n)} \frac{\Gamma(\psi / \phi + k)}{\Gamma(\psi / \phi)} \phi^{k} \cdot$$
$$\cdot \prod_{g = 1}^{k} \frac{\Gamma(n_{g} - \phi)}{\Gamma(1 - \phi)}$$
Prior distribution on $C_{k}$
Independently classify each column $j$ with probabilities $\gamma_{j} = (\gamma_{j1}, \ldots, \gamma_{ju}, \ldots, \gamma_{jv})^{\prime}$
If $j$ is classified as $u$, independently sample within cluster $g$ with probabilities $\omega_{jg}^{u} = (\omega_{jg1}^{u}, \ldots, \omega_{jgl}^{u}, \ldots, \omega_{jgs_{u}}^{u})^{\prime}$
Joint distribution is then
$$p(C_{k} | k) = \prod_{j = 1}^{m} \prod_{g = 1}^{k} \prod_{u = 1}^{v} \prod_{l = 1}^{s_{u}} \left(\gamma_{ju}^{\frac{1}{k}} \omega_{jgl}^{u}\right)^{c_{jgl}^{u}}$$
Bayesian bi-clustering model
$$p(k, R_{k}, C_{k} | X) \propto p(k, R_{k}) \prod_{j = 1}^{m} \prod_{g = 1}^{k} \prod_{u = 1}^{v} \prod_{l = 1}^{s_{u}} \left(\gamma_{ju}^{\frac{1}{k}} \omega_{jgl}^{u} p_{jgl}^{u}(x_{gj})\right)^{c_{jgl}^{u}}$$
where $X$ is the categorical data encoded as a presence/absence binary matrix and
$$p_{jgl}^{u}(x_{gj}) = p(x_{gj} | c_{jgl}^{u} = 1) = \int p(\theta | c_{jgl}^{u} = 1) \prod_{i \in g} p(x_{ij} | \theta) d\theta$$
Posterior distribution
We can show that
$$p(X | k, R_{k}) = \prod_{j = 1}^{m} \sum_{u = 1}^{v} \gamma_{ju} \prod_{g = 1}^{k} \sum_{l = 1}^{s_{u}} \omega_{jgl}^{u} p_{jgl}^{u}(x_{gj})$$
$$p(C_{k} | k, R_{k}, X) = \prod_{j = 1}^{m} \prod_{u = 1}^{v} \left(\frac{\gamma_{ju} \prod\limits_{h = 1}^{k} \sum\limits_{t = 1}^{s_{u}} \omega_{jht}^{u} p_{jht}^{u}} {\sum\limits_{a = 1}^{s_{u}} \gamma_{ja} \prod\limits_{h = 1}^{k} \sum\limits_{t = 1}^{s_{u}} \omega_{jht}^{a} p_{jht}^{a}} \right)^{\frac{c_{j..}^{u}}{k}} \prod_{g = 1}^{k} \prod_{l = 1}^{s_{u}} \left[\frac{\omega_{jgl}^{u} p_{jgl}^{u}}{\sum\limits_{t = 1}^{s_{u}} \omega_{jgt}^{u} p_{jgt}^{u}}\right]^{c_{jgl}^{u}}$$
from which we can implement merge-split Metropolis-Hastings and Gibbs sampling algorithms
Rotavirus A VP4 protein (clustered data)
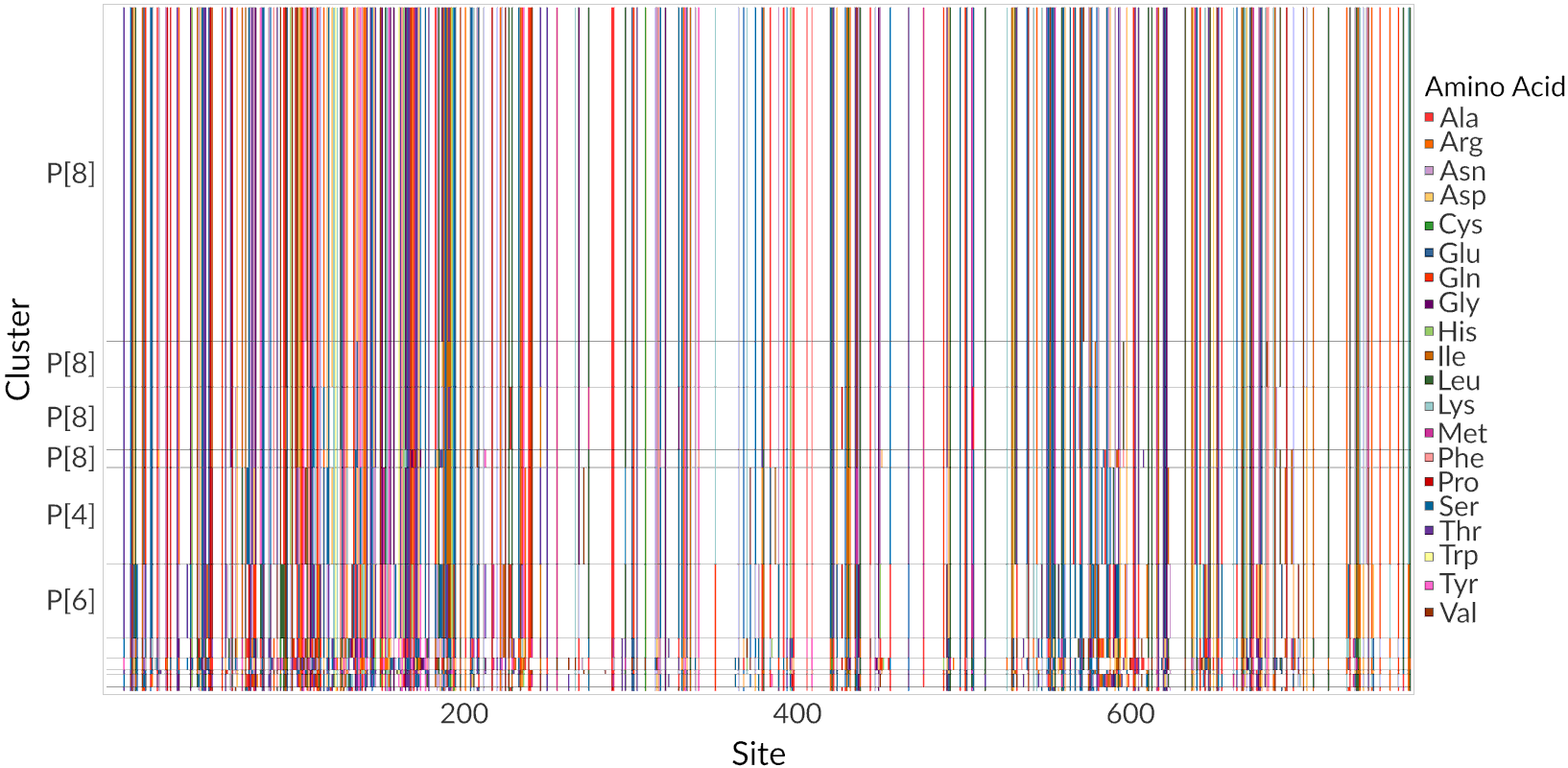
Rotavirus A VP4 protein (Row clusters)
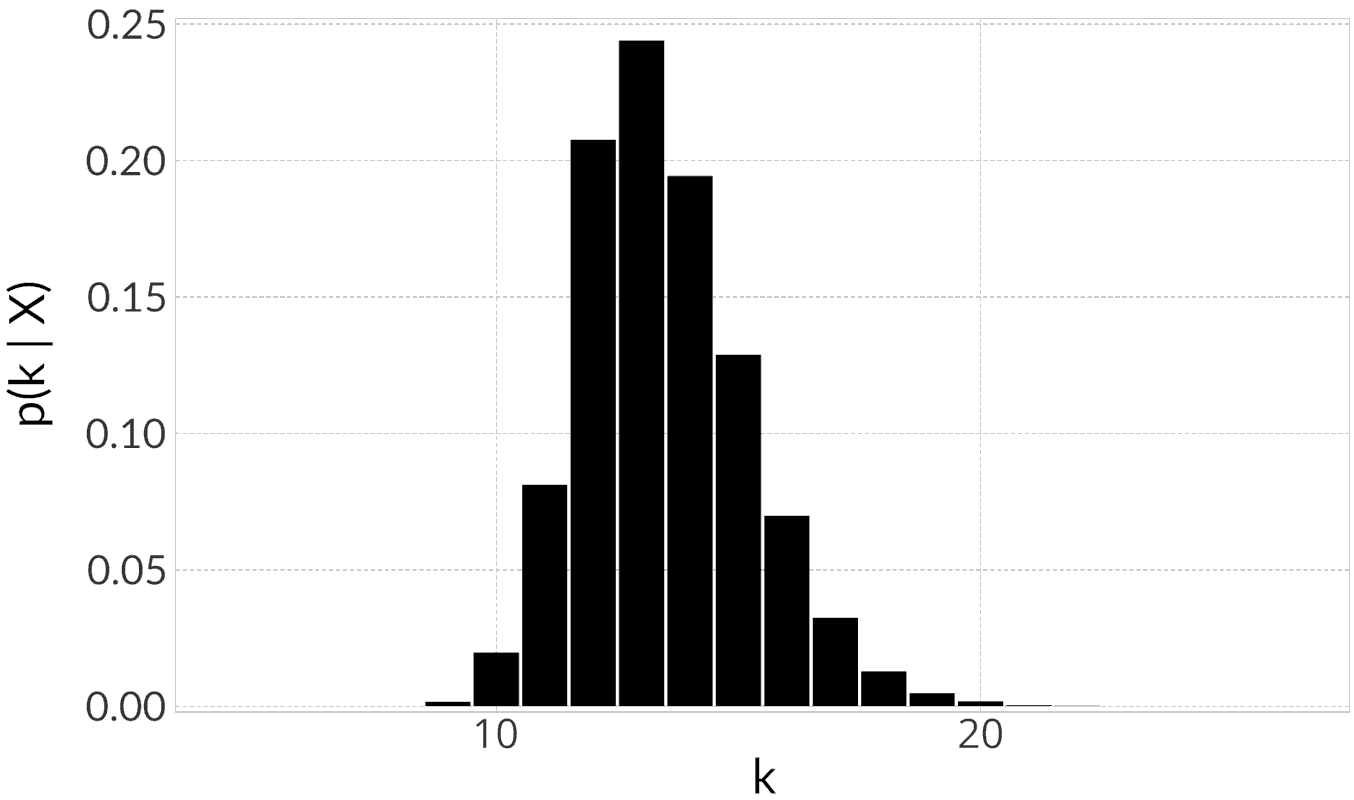
|
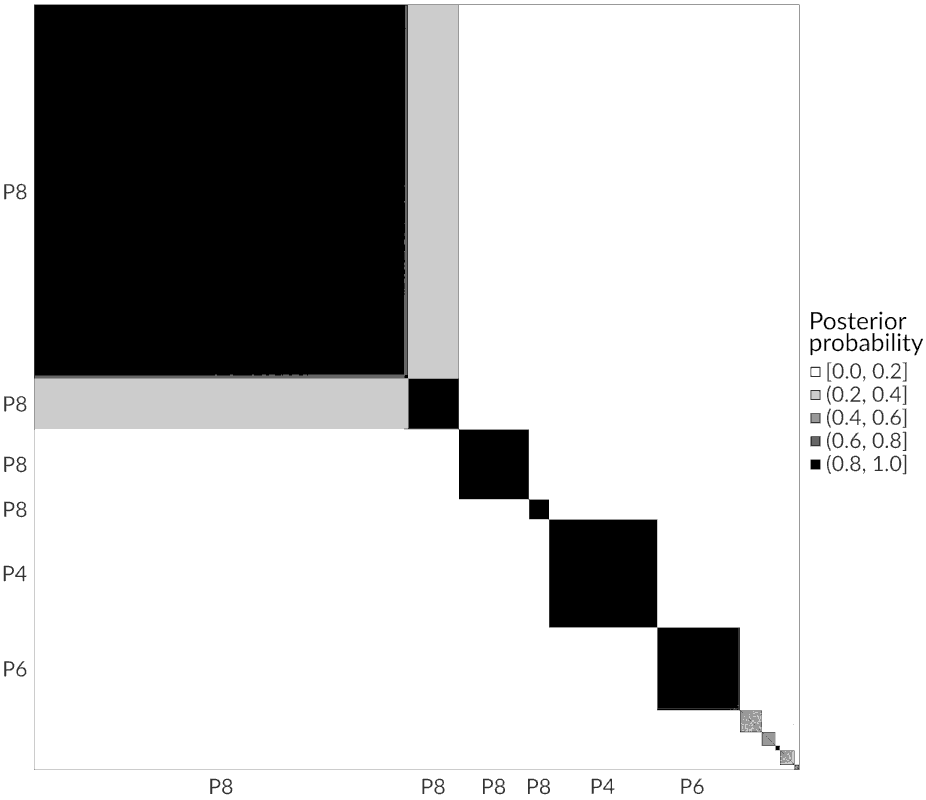
|
Rotavirus A VP4 protein (Column classification)
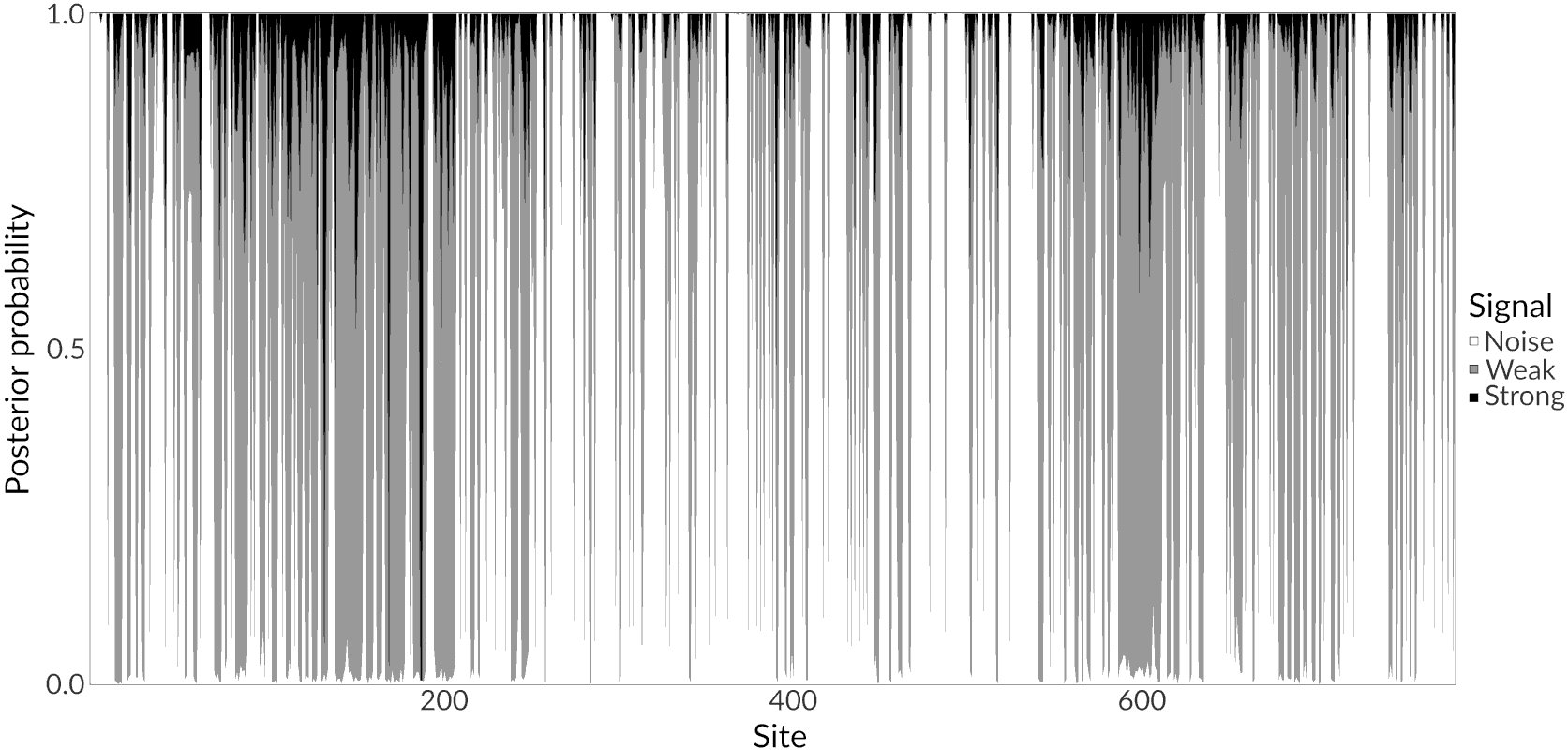
Conclusions
- Model is general enough to be (theoretically) extended to other settings
- Cluster analysis is a computationally hard problem
- MCMC algorithms exist but don’t scale well with the sample size
- Implementation of our model is available as a Julia package: https://github.com/albertopessia/Kpax3.jl